Impact of MLL::AF9 Gene Rearrangement on Survival of Acute Myeloid Leukaemia Patients: An Insight into Pakistani Population
By Muhammad Tariq1,2, Sadaf Shahab2, Javeria Rauf Saeed3, Zeeshan Hussain4, Uzma Zaidi2, Tasneem Farzana2, Sultan Ahmad5Affiliations
doi: 10.29271/jcpsp.2024.04.424ABSTRACT
Objective: To ascertain the frequency of the MLL::AF9 gene rearrangement and its association with survival in Pakistani patients suffering from acute myeloid leukaemia (AML).
Study Design: Analytical study.
Place and Duration of the Study: Department of Haematology, National Institute of Blood Diseases and Bone Marrow Transplantation, Karachi, Pakistan, from 2015 to 2020.
Methodology: Patients without a history of past AML chemotherapy, aged from 10 to 75 years, were included. Individuals with metastatic cancer, chronic myeloid leukaemia, or other haematological conditions were excluded. Identifying the MLL::AF9 gene involved RNA extraction, cDNA synthesis, and Real-time PCR amplification. The Chi-square test was used to examine the relationship between survival and the MLL::AF9 mutation. A Welch two-sample t-test was used to evaluate survival days depending on the MLL::AF9 gene rearrangement, while ANOVA was used to analyse survival days across various death statuses.
Results: The mean age of 130 patients was 36.65 ± 13.01 years, with 64.62% being males. The most common leukaemia type was AML-M2 (n = 32, 24.62%). During the study follow-up, 22.31% were still alive, 40.77% died, and the status of 36.92% were unknown. MLL::AF9 gene rearrangement was present in 11.54%. The group with MLL::AF9 gene rearrangement had significantly longer mean ‘survival days’ (1,542.33 ± 926.07) compared to the group without the gene rearrangement (206.42 ± 359.57, p <0.001).
Conclusion: MLL-AF9 mutation was present in 11.54%. Age and MLL::AF9 gene rearrangement were significant predictors of survival in leukaemia patients.
Key Words: Acute myeloid leukaemia, MLL::AF9, Gene rearrangement, Survival.
INTRODUCTION
Acute myeloid leukaemia (AML) is a rapidly progressing blood malignancy characterised by aberrant stem or progenitor cell proliferation inside the bloodstream.1,2 Despite extensive research efforts over many years, AML continues to have a high mortality rate (less than 30% survival rate after 5 years) and a tendency to recur, with limited treatment choices available.
AML is triggered and maintained by a small portion of leukaemia stem cells (LSCs) that possess unique biological characteristics compared to the majority of tumour cells. These LSCs exhibit varying sensitivity to chemotherapy and play a significant role in treatment resistance and disease relapse.3
Modifications in the mixed lineage leukaemia (MLL) gene are linked to 10% of cases of acute leukaemia, encompassing both acute lymphoblastic leukaemia (ALL) and acute myeloid leukaemia (AML).4 The MLL gene, alternatively referred to as KMT2A, participates in chromosomal rearrangements in 5-10% of acute leukaemias. These rearrangements play a crucial role in determining therapy stratification, assessing minimal residual disease, and targeted therapies.5 Though 79 different MLL translocation partner genes have been identified, a few specific rearrangements make up the bulk of MLL aberrations in AML.6,7 AML with MLL gene modifications display clinical and immunological diversity, leading to a range of immunophenotypes. The outlook for AML is typically grim and hinges on factors such as the precise translocation, phenotype, and the patient's age. The t(9;11) (p22;q23) translocation is linked to notably extended patient survival rates, whereas none of the 11q23 abnormalities bodes well for prognosis.8 The MLL::AF9 (Mixed-Lineage Leukaemia-AF9) gene fusion is a genetic abnormality associated with subtypes of AML. The MLL gene plays a crucial role in overseeing the expression of genes vital for the regular development of blood cells.9 However, when a chromosomal translocation occurs between the MLL gene on chromosome 11 and a partner gene, such as AF9 on chromosome 9, it creates the MLL::AF9 fusion gene. This fusion gene produces a protein that disrupts the normal regulation of gene expression, leading to the development of AML.10
MLL::AF9 is most commonly found in a subtype of AML known as M5 or acute monocytic leukaemia, but it can also occur in other subtypes.11 Compared to other types of AML, the MLL::AF9 gene fusion is linked to a prognosis ranging from poor to moderate in AML. Patients with this specific genetic alteration tend to have a high response rate in comparison to other types, given standard chemotherapy. Nonetheless, the outlook can fluctuate based on several factors, such as the patient's age, the existence of additional genetic mutations, and the response to treatment.4
The MLL::AF9 fusion protein interacts with various cellular components involved in gene regulation, including proteins that modify histones (proteins that help package DNA), leading to dysregulated gene expression.9 This dysregulation promotes the uncontrolled growth and proliferation of leukaemic cells. MLL::AF9 also affects the self-renewal capacity of leukaemic stem cells, contributing to the persistence and recurrence of the disease.10
In the MLL gene fusion, approximately 100 partner genes have been documented. However, data related to clinical outcomes for the 11q23-rearranged subgroups is scarce, mainly because this subset constitutes a small portion, making meaningful analysis a challenge. According to the literature, patients with t(9;11)(p22;q23) generally exhibit a more favourable prognosis compared to those with other MLL translocations.6
There is a notable gap in research regarding the specific influence of the MLL::AF9 gene rearrangement on survival time in individuals with newly diagnosed AML. Researchers can learn more about the underlying processes of the disease by investigating its genetic background and concentrating on the MLL::AF9 gene rearrangement. This knowledge might help identify possible therapeutic targets, which would improve therapy, lessen side effects and improve patient care. The objective of this study was to ascertain the frequency of the MLL::AF9 gene rearrangement and its association with survival in Pakistani patients suffering from AML.
METHODOLOGY
This analytical cross-sectional study was conducted on 130 AML patients via non-probability consecutive sampling technique from 2015 to 2020. Approval for this study was granted by the institutional review board of the National Institute of Blood Diseases and Bone Marrow Transplantation (NIBD and BMT) under reference NIBD/RD-162/17-2014. Following an extensive elucidation of the study, written informed consent was acquired from every participant and the data of the patients was retrieved from the medical record maintained at NIBD and BMT. They were given assurance that all gathered data would be treated with strict confidentiality and anonymity. Written informed consent was taken from each patient.
Newly diagnosed male and female patients, aged between 10 and 75 years, with no history of previous chemotherapy, were included in the study. Patients with chronic myeloid leukaemia evolving into AML, any other malignancy, or other coexisting haematological disorders were excluded from the study.
AML patients were confirmed through bone marrow biopsy and immunohistochemistry, following the diagnostic criteria of the French-American-British (FAB) classification and World Health Organization (WHO) classification of haematolymphoid malignancies. A detailed history was taken, which included information about age, gender, clinical features, complete blood count (CBC), bone marrow aspirate, and biopsy reports. Both DNA and RNA were extracted from the collected blood or bone marrow samples. All the selected patients were followed up for a maximum period of 8 years.
The RNA extraction was carried out using the Triazole method as described.12 The complementary DNA (cDNA) was prepared from RNA using the Affymetrix First-strand cDNA synthesis kit, following the manufacturer's instructions. The PCR master mix was prepared using the Ana Gene Biotech real-time PCR kit, according to the manufacturer's recommendations. To prepare the PCR mix, 2 μl of patient cDNA was added to 10 μl of qPCR mix (2x), followed by the addition of 5 μl of MLL/AF9 probe/primer mix and 3 μl of nuclease-free water into a PCR tube. The mixture was gently pipetted up and down several times to ensure thorough mixing. Finally, the PCR strips were placed in the real-time instrument Roter Gene Q for further analysis.
FAM green channel was used for translocation detection while HEX yellow channel for internal control. After completion of cycles, the fusion transcripts were analysed in the FAM channel and internal control (ABL gene) in the HEX channel. Only those sample results were considered valid whose internal control (ABL gene) Ct values were before 30 cycles. The signals of translocation with Ct values below 35 for FAM signal and amplification curve with exponential growth were considered as true positive.
Statistical analyses were conducted using the R statistical software package, version 1.3.2. Mean and standard deviation (SD) were computed for numerical data, while categorical variables were presented using percentages and frequencies. A Kaplan-Meier plot was generated to illustrate the survival rates for individuals with and without the MLL::AF9 gene mutation. The association between MLL::AF9 gene rearrangement and death status was determined using Chi-square test. ANOVA was conducted to compare survival days across different death statuses. Furthermore, Welch two-sample t-test was applied to assess the variation in survival days depending on the presence of the MLL::AF9 gene rearrangement. Additionally, Cox proportional regression analysis was conducted, with survival days acting as the dependent variable while MLL::AF9 gene rearrangement, age, and gender as independent variables. A significance level of p <0.05 was deemed statistically significant.
RESULTS
A total of 130 leukaemia patients were included. Their mean age was 36.65 ± 13.01 years. There were 84 (64.62%) males and 46 (35.38%) females. The mean haemoglobin level among these patients was 8.08 ± 2.04 gm/dl. Mean leukocyte count was observed as 45.56 ± 73.58 x 109 /L, while the mean percentage of blasts in the blood was 53.15 ± 28.50%. The platelet count showed an average value of 69.96 ± 75.22 x 109 /L. The mean survival time for these patients was found 360.56 ± 625.23.
The most common type of leukaemia was AML-M2 (n = 32, 24.62%) followed by AML (n = 31, 23.85%) and then AML-M1 (n = 25, 19.23%). The least frequent was AML-M7 found only in 2 patients (1.54%). Throughout the study follow-up, 29 patients (22.31%) were alive and 53 patients (40.77%) died, while for 48 (36.92%) of the patients the status was unknown. MLL::AF9 gene rearrangement was found in 15 patients (11.54%).
The Chi-square test p-value indicated that there was no significant association between the MLL::AF9 gene rearrangement and survival statuses (p = 0.204). The data indicated that the majority of patients in each group were negative for MLL::AF9 gene rearrangement. Specifically, 86.21% of patients in the alive group, 94.34% in the expired group, and 83.33% in the unknown group were negative for MLL::AF9 gene rearrangement.
The ANOVA test suggested that there were significant differences in survival days between the three groups (p <0.001). The mean survival days for the alive group (mean = 644.72 ± 958.68) was significantly higher compared to the expired group (153.09 ± 243.74) and the unknown group (417.96 ± 598.28, p <0.001 for both comparisons). However, there was no significant difference between the unknown group and the alive group (p = 0.11), but there was a significant difference between the expired group and the unknown group (p = 0.028).
Table I presents the results of a Welch two-sample t-test that was used to compare the mean survival days between two groups based on the presence or absence of the MLL::AF9 gene rearrangement. The p-value, which is less than 0.001, suggested that there was a significant difference in the mean survival days between the two groups. The group with the positive mutation had a significantly longer mean survival days (mean = 1,542.33 ± 926.07) compared to the group without mutation (206.42 ± 359.57). Kaplan Meir graph (Figure 1) showed higher survival probability in AML patients with MLL::AF9 gene rearrangement.
The MLL::AF9 gene rearrangement is associated with longer survival time (coef = -1.724). The older age is associated with lower survival time (coef = 0.023). While male gender is also associated with shorter survival time (coef = 0.629). The hazard is 0.178 times lower for individuals with the MLL- AF9 mutation compared to those without the mutation (i.e., the hazard is 82% lower). Similarly, the hazard of the event of interest is 1.02 times higher for each one-year increase in age and 1.88 times higher for males compared to females. Age and MLL::AF9 gene rearrangement were significant predictors of survival in leukaemia patients as shown in Table II.
Table I: Comparison of survival days and MLL::AF9 gene rearrangement of leukaemia patients.
|
Characteristic |
MLL::AF9 gene rearrangement |
p-value* |
|
|
Absent (Negative) N = 115 |
Present (Positive) N = 15 |
||
|
Survival days, Mean ± SD |
206.42 ± 359.57 |
1,542.33 ± 926.07 |
<0.001 |
|
*Welch two sample t-test N = no. of Patients. |
|||
Table II: Cox proportional regression analysis for survival days for leukaemia patients.
|
Survival days |
coef |
Hazard ratio [exp(coef)] |
se(coef) |
z |
p-value |
|
MLL::AF9 gene rearrangement (present) |
-1.72496 |
0.17818 |
0.64260 |
-2.684 |
0.00727 |
|
Age |
0.02344 |
1.02371 |
0.01003 |
2.336 |
0.01949 |
|
Gender (male) |
0.62976 |
1.87716 |
0.33360 |
1.888 |
0.05906 |
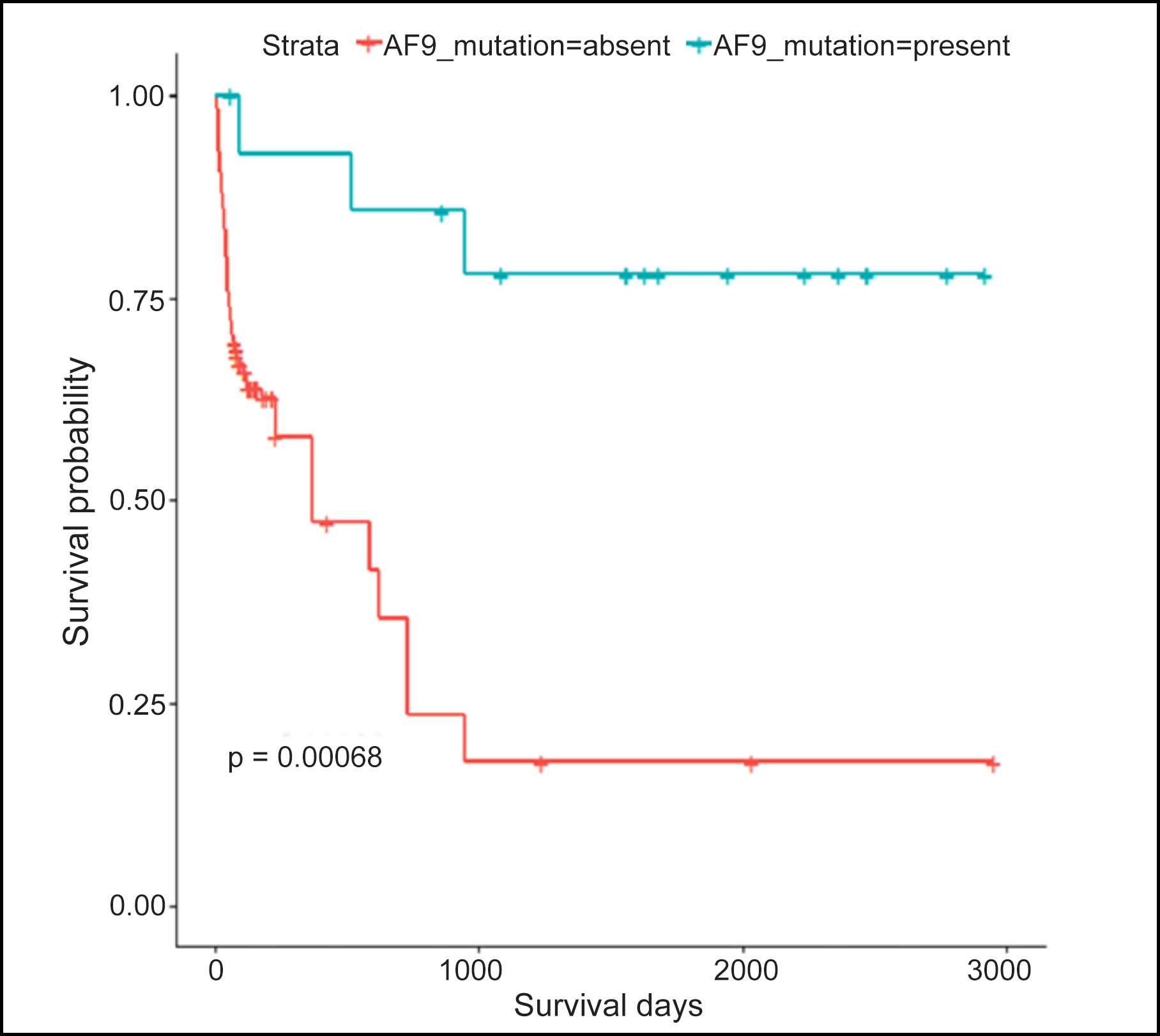 Figure 1: Kaplan Meir for survival days and MLL::AF9 gene rearrangement.
Figure 1: Kaplan Meir for survival days and MLL::AF9 gene rearrangement.
DISCUSSION
This study explored the impact of MLL::AF9 gene mutation on the survival of AML patients. Out of total 130 participants, data on survival status at the end of the study were available for 82 participants. AML was chosen as the target for several reasons, primarily because it is more prevalent in adults and is associated with a less favourable outcome. Adults typically demonstrate better compliance with follow-up.
The frequency of MLL::AF9 gene mutation in survived leukaemia patients was found to be more prevalent than in the patients who died, showing some protective effect against the deadly disease, but the study found no significant association between MLL::AF9 gene rearrangement and survival statuses (p = 0.204). It might be due to 48 cases with unknown status of survival mainly due to loss to follow-up. The group with the mutation had significantly longer mean survival days compared to the group without the mutation.
Subtypes of leukaemia in the current study were AML-M2 (n = 32, 24.62%) followed by AML (n = 31, 23.85%) and then AML-M1 (n = 25, 19.23%). The least frequent type of leukaemia is AML-M7 with 1.54 %. Throughout the study follow-up, 29 (22.31%) were still alive and 53 (40.77%) died while for 48 (36.92%) the status was unknown. Variation in subtypes of leukaemia is known to vary widely worldwide due to geographic variation. Literature reports different sub-classes of AML with different prevalence frequencies, it might be due to age, ethnicity, race, and geographic variation, which suggests that different subtypes may have diverse aetiologic factors.13 For that reason, a detailed comprehensive evaluation of leukaemia subtype patterns is necessary globally. The present study shows a higher prevalence of AML in males. Likewise, the incidence of AML in males is higher as compared to females in a study conducted by Stabellini et al., along with a notable predominance of AML in males, overall survival in females is also good (lower risk of death).14
The study showed the average age of leukaemia patients at the time of diagnosis as 36.65 ±13.01 years while the mean haemoglobin level, platelets counts were low with high WBC count. Other studies conducted in Pakistan and India respectively validated these results.15,16 The data showed the mean survival of AML patients as 360.56 ± 625.23 days which is also supported by Kakepoto et al.17 The main emphasis of the current project was to report frequency of MLL::AF9 gene rearrangement in AML cases, which was detected in 15 patients, which is almost similar to a study conducted in China.18 In this study, among the patients who underwent partner gene detection (28.1%) were positive for MLL::AF9 detection, likewise reported 272 adult AML patients with MLL gene rearrangement, and 71 patients (26.1%).
With MLL::AF9 t(9;11) (p22;q23) mutation.19 Literature shows that MLL::AF9 positive AML patients have a healthier remission rate and survival than other sub-types of AML patients with other MLL gene rearrangements.20
In the present study, the mean blast percentage in AML patients was 53.15 ± 28.50%, which is consistent with previous literature findings.21 Significant differences were observed in the association between MLL::AF9 gene rearrangement and patient death status, as shown by the results of this study. These findings align with a study conducted by Scholl et al.22 which also reported a lower death ratio among AML patients with the MLL::AF9 gene mutation compared to those without it. Additionally, a significant difference in survival days among leukaemia patients with and without MLL::AF9 gene rearrangement was identified. These results are also consistent with the study conducted by Stubbs et al.23
Despite the valuable findings of this study, several limitations need to be acknowledged. First, the small size did not allow the authors to establish a relationship between the MLL::AF9 gene mutation and survival outcomes in AML patients. It is important to consider that other confounding factors could also be influencing the observed results. Secondly, the presence of cases with unknown survival status due to loss to follow-up introduces a significant limitation. This missing data can potentially be biased and have impacted the findings. The unknown status of these cases prevents a comprehensive analysis of the entire patient cohort and may affect the statistical significance of the observed associations. Moreover, it is essential to recognise that survival outcomes in AML are influenced by various factors other than the MLL::AF9 gene rearrangement. Other genetic abnormalities, treatment protocols, patient characteristics, and coexisting health conditions can significantly impact patient survival. The study did not account for the other potential confounding effects of these factors, which limits the ability to attribute the observed differences solely to the MLL::AF9 gene rearrangement.
CONCLUSION
MLL- AF9 mutation was present in 11.54%. Age and MLL::AF9 gene rearrangement were significant predictors of survival in leukaemia patients while this study provides valuable insights for protective effect of MLL::AF9 gene mutation against mortality in AML patients, it is important to interpret the findings within the context of its limitations.
ETHICAL APPROVAL:
Approval for this study was granted by the institutional review board of the National Institute of Blood Diseases and Bone Marrow Transplantation (NIBD & BMT) under reference NIBD/RD-162/17-2014, before initiation of the research work.
PATIENTS’ CONSENT:
Written informed consent was acquired from every participant before the collection of samples and data. They were given assurance that all gathered data would be treated with strict confidentiality and anonymity and would be used only for research purposes.
COMPETING INTEREST:
The authors declared that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AUTHORS’ CONTRIBUTION:
MT: Write-up, design, conception, data acquisition.
SS: Design, conception, write up, data acquisition.
JR, TF, UZ: Data acquisition, data analysis, interpretation.
ZH, SA: Review the literature, interpretation.
All authors read and approved the final version of manuscript to be published.
REFERENCES
- Tallman MS, Wang ES, Altman JK, Appelbaum FR, Bhatt VR, Bixby D, et al. Acute myeloid leukaemia, version 3.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw 2019; 17(6):721-49. doi: 10.6004/jnccn.2019. 0028.
- Khwaja A, Bjorkholm M, Gale RE, Levine RL, Jordan CT, Ehninger G, et al. Acute myeloid leukaemia. Nat Rev Dis Primers 2016; 2:16010. doi: 10.1038/nrdp.2016.10.
- Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell 2014; 14(3):275-91. doi: 10.1016/j.stem. 2014.02.006.
- Winters AC, Bernt KM. MLL-rearranged leukemias-an update on science and clinical approaches. Front Pediatr 2017; 5:4. doi: 10.3389/fped.2017.00004.
- Issa GC, Ravandi F, DiNardo CD, Jabbour E, Kantarjian HM, Andreeff M. Therapeutic implications of menin inhibition in acute leukemias. Leukemia 2021; 35(9):2482-495. doi: 10.1038/s41375-021-01309-y.
- Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: Results of an international retrospective study. Blood 2009; 114(12):2489-96. doi: 10.1182/blood-2009- 04-215152.
- Patel U, Luthra R, Medeiros LJ, Patel KP. Diagnostic, prognostic, and predictive utility of recurrent somatic mutations in myeloid neoplasms. Clin Lymphoma Myeloma Leuk 2017; 17S:S62-74. doi: 10.1016/j.clml.2017.02.015.
- Ilencikova D, Kolenova A. MLL gene alterations in acute myeloid. Oncogene and cancer- From bench to clinic. 2013;225. doi:10.5772/55141.
- Lomov N, Zerkalenkova E, Lebedeva S, Viushkov V, Rubtsov MA. Cytogenetic and molecular genetic methods for chromosomal translocations detection with reference to the KMT2A/MLL gene. Crit Rev Clin Lab Sci 2021; 58(3): 180-206. doi: 10.1080/10408363.2020.1844135.
- El Chaer F, Keng M, Ballen KK. MLL-Rearranged acute lymphoblastic leukemia. Curr Hematol Malig Rep 2020; 15(2):83-89. doi: 10.1007/s11899-020-00582-5.
- Lewis AH, Bridges CS, Moorshead DN, Chen TJ, Du W, Zorman B, et al. Krüppel-like factor 4 supports the expansion of leukemia stem cells in MLL-AF9-driven acute myeloid leukemia. Stem Cells 2022; 40(8):736-50. doi: 10.1093/stmcls/sxac033.
- Rio DC, Ares M Jr, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc 2010; 2010(6):pdb.prot5439. doi: 10.1101/pdb.prot5439.
- Kulshrestha R, Sah SP. Pattern of occurrence of leukemia at a teaching hospital in eastern region of Nepal - A six year study. JNMA J Nepal Med Assoc 2009; 48(173):35-40. doi:10.31729/jnma.194.
- Stabellini N, Tomlinson B, Cullen J, Shanahan J, Waite K, Montero AJ, et al. Sex differences in adults with acute myeloid leukemia and the impact of sex on overall survival. Cancer Med 2023; 12(6):6711-21. doi: 10.1002/cam4.5461.
- Kakepoto GN, Burney IA, Zaki S, Adil SN, Khurshid M. Long-term outcomes of acute myeloid leukemia in adults in Pakistan. J Pak Med Assoc 2002; 52(10):482-6.
- Kumar M, Gaur V, Chowdhry M, Raj NM, Sharma P. Hemoglobin level and karyotype status are independent prognostic parameters for acute myeloid leukemia: Pilot study of 244 patients. Int J Med Res Health Sci 2019; 8(6):135-42.
- Jenkins N, Blanshard LA, Stone M, Verburgh E, Oosthuizen J, Shires K. Cytogenetically normal acute myeloid leukaemia at a single centre in South Africa. Hematol Oncol Stem Cell Ther 2023; 16(4):397-406. doi: 10.56875/2589-0646.1087.
- Liu K, Li Y, Qiu S, Zhou C, Wei S, Lin D, et al. Efficacy of combination of venetoclax with azacitidine or chemo-therapy in refractory/relapse acute leukemias of ambiguous lineage, not otherwise specified. Exp Hematol Oncol 2021; 10(1):46. doi: 10.1186/s40164-021-00239-w.
- Meyer C, Hofmann J, Burmeister T, Gröger D, Park TS, Emerenciano M, et al. The MLL recombinome of acute leukemias in 2013. Leukemia 2013; 27(11):2165-76. doi: 10.1038/leu.2013.135.
- Krauter J, Wagner K, Schäfer I, Marschalek R, Meyer C, Heil G, et al. Prognostic factors in adult patients up to 60 years old with acute myeloid leukemia and translocations of chromosome band 11q23: Individual patient data-based meta-analysis of the German acute myeloid leukemia intergroup. J Clin Oncol 2009; 27(18):3000-6. doi: 10.1200/ JCO.2008.16.7981.
- DiNardo CD, Garcia‐Manero G, Pierce S, Nazha A, Bueso‐Ramos C, Jabbour E, et al. Interactions and relevance of blast percentage and treatment strategy among younger and older patients with acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). Am J Hematol 2016; 91(2):227-32. doi: 10.1002/ajh.24252.
- Konuma T, Mizuno S, Kondo T, Yamaguchi H, Fukuda T, Uchida N, et al. Allogeneic hematopoietic cell trans-plantation in adult acute myeloid leukemia with 11q23 abnormality: A retrospective study of the adult acute myeloid leukemia working group of the Japan Society for Hematopoietic Cell Transplantation (JSHCT). Ann Hematol 2018; 97(11):2173-83. doi: 10.1007/s00277-018-3419-1.
- Stubbs MC, Kim YM, Krivtsov AV, Wright RD, Feng Z, Agarwal J, et al. MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: Development of a model for rapid therapeutic assessment. Leukemia 2008; 22(1):66-77. doi: 10.1038/sj.leu.2404951.